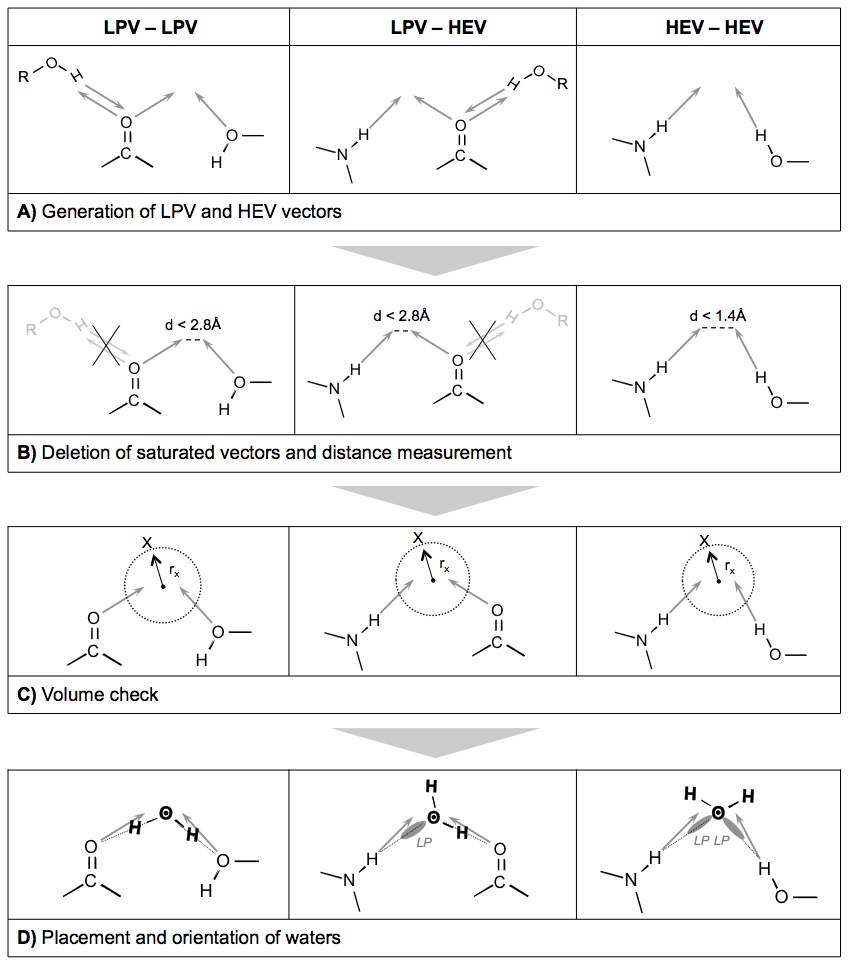
In AcquaAlta explicit water molecules are generated at the ligand–protein interface.
The underlying algorithm relies on preferred positions and orientations of water molecules as extracted from structural information collected from the Cambridge Structural Database — CSD.
Specifically, we searched for water molecules interacting with generic functional groups of small organic molecules.
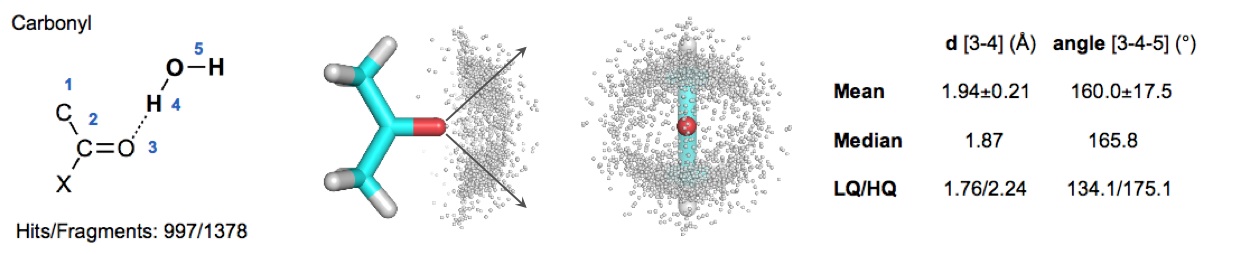
Example of CSD query and results from the CSD search
To establish a hydration-propensity ranking, water interaction energies were obtained from ab initio calculations on hydrated functional groups. In AcquaAlta, water molecules bridging interactions between ligand and protein partners are generated considering the hydration propensities of the involved functional groups and aromatic moieties.
The following flowchart explains the underlying procedure used to generate water molecules.